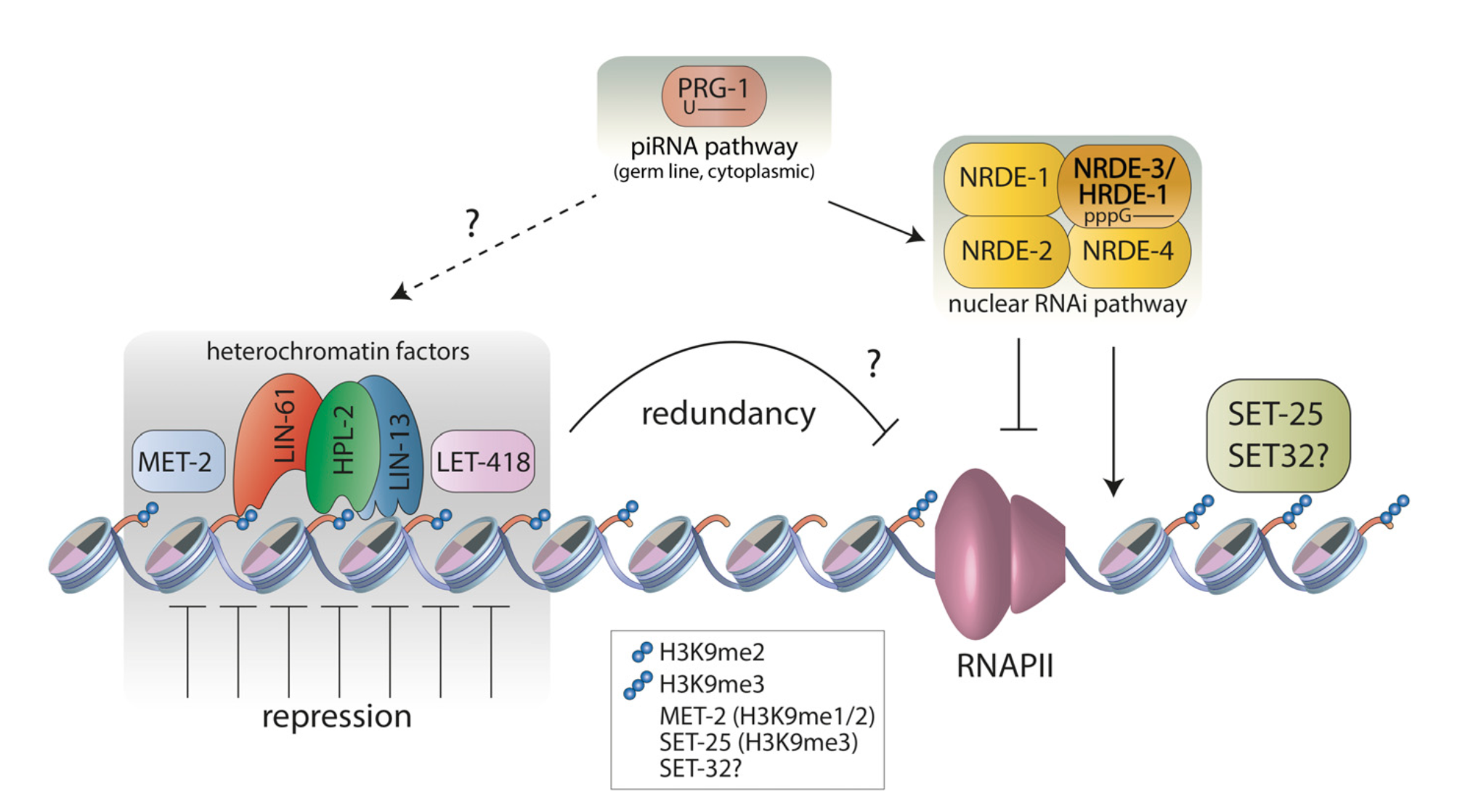
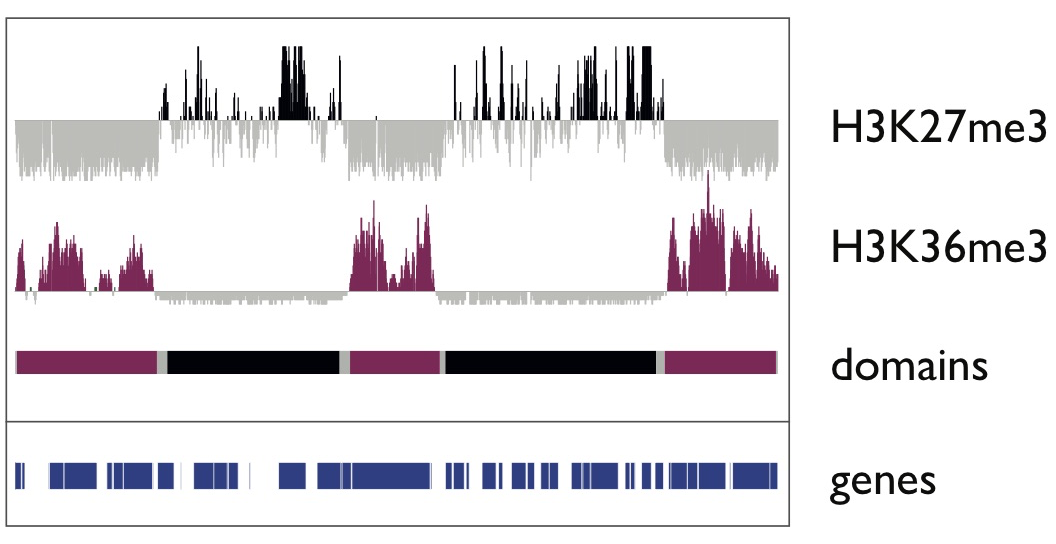
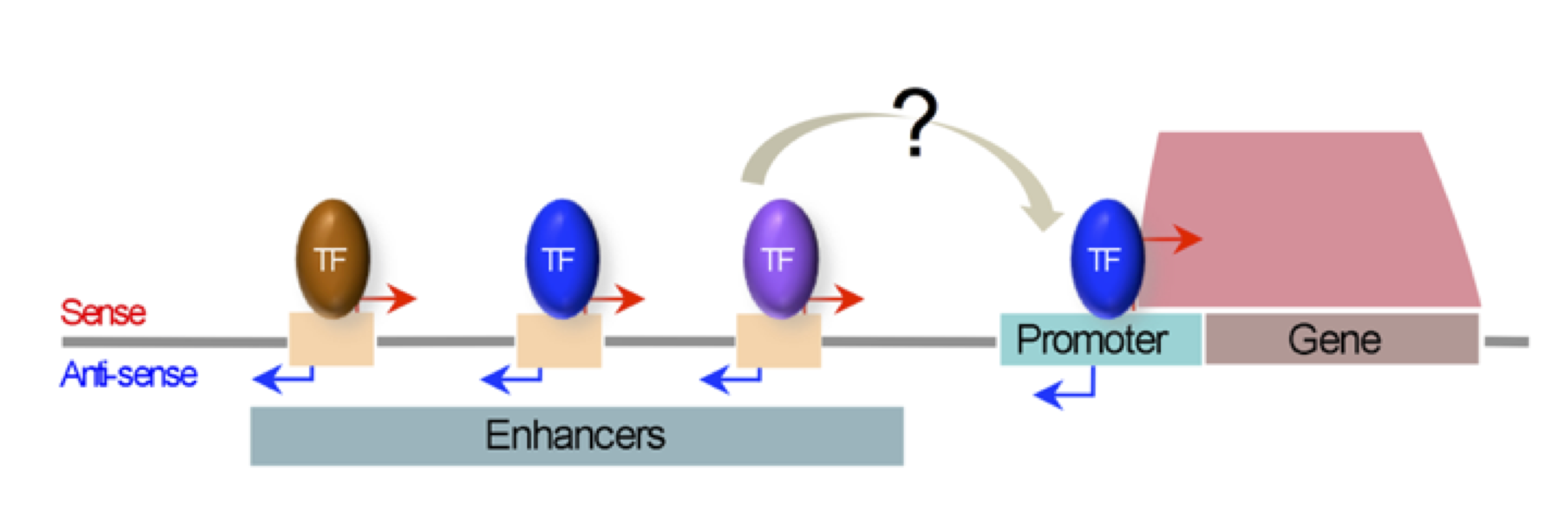
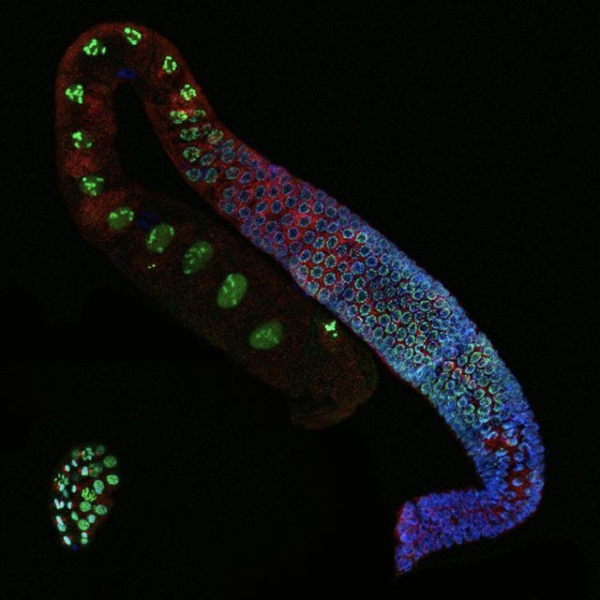
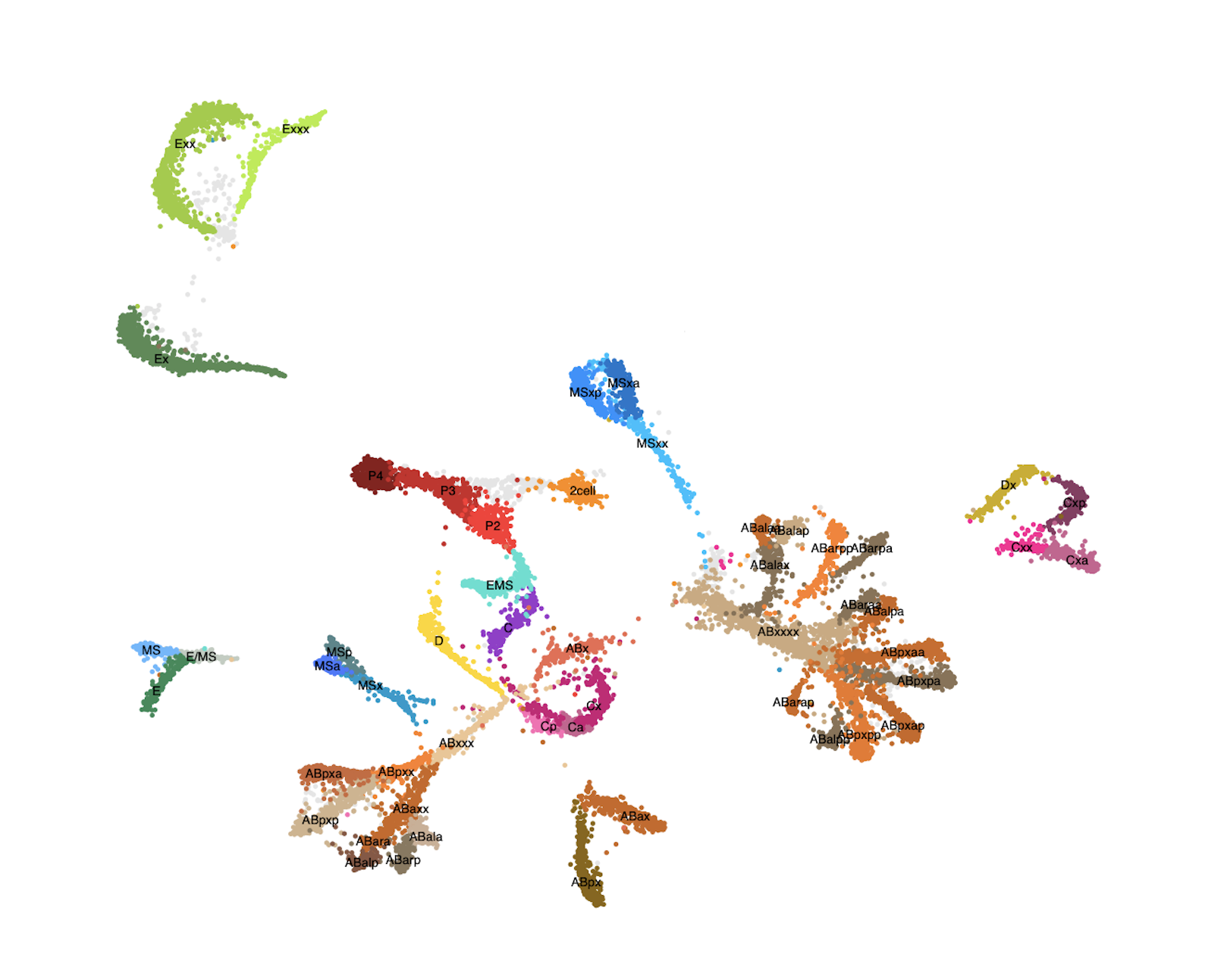A fundamental property of a single animal genome sequence is the ability to give rise to diverse cell types. The regulation of chromatin establishes gene expression programmes that drive cellular identity, and dysregulation can cause disease. We study how chromatin and gene expression are regulated across developmental trajectories to ultimately generate and maintain the correct cell types. We use the C. elegans animal model system, in which we leverage the awesome powers of genomic technologies such as lineage-resolved single-celled multiomics and diverse high-throughput chromatin assays, together with RNAi screening, genome-editing, and super-resolution microscopy.