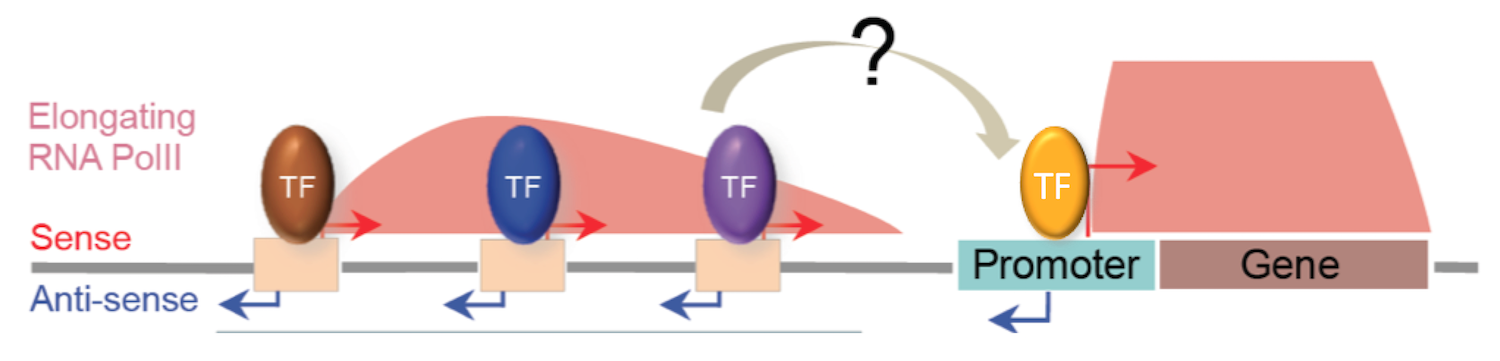
To regulate transcription, chromatin factors must impact the activities of promoters and enhancers, for example through altering their accessibility. Enhancers are classically bound by sequence-specific transcription factors that either activate or repress transcription from nearby core promoters. The interactions and regulatory events that occur between enhancers and promoters are poorly understood.
In previous work, we determined the genome-wide landscape of RNA Polymerase II transcription initiation and elongation in C. elegans (Chen et al, 2013). Unexpectedly, most transcription initiation events occurred at enhancers, generating long nuclear specific transcripts. Transcription has been also been observed at mammalian enhancer regions, but it is not well-characterized and its function is unclear. We also identified other types of non-coding RNAs, including hundreds of transcripts antisense to protein coding genes and the promoters that drive them. Enhancer and antisense ncRNAs could have regulatory roles, or the process of transcription could act to generate regions of open chromatin in order to enhance accessibility. We are functionally analysing enhancers and promoters and the roles of non-coding transcription using transgenic and gene targeting approaches. We are also systematically defining regulatory elements genome-wide and through development, using DNAse I, micrococcal nuclease and ATAC-seq.